The P. knowlesi parasite is transmitted from monkeys, including long-tailed macaques (pictured), to humans via mosquitoes and can cause severe malaria. UNIMAS researchers are investigating subpopulations of the parasite.
This story is featured in the Asia Research News 2021 magazine. Read in ISSUU (above) or full text and images below.
Malaria caused by the monkey parasite Plasmodium knowlesi is a serious public health concern in Malaysian Borneo, and cases are on the rise. Researchers from Universiti Malaysia Sarawak (UNIMAS), together with collaborators from the London School of Hygiene & Tropical Medicine, have developed a simple tool to track different subpopulations of this parasite species, revealing new insights into the transmission of the disease.
The P. knowlesi parasite is transmitted from monkeys to humans via mosquitoes and can cause severe malaria, which can be fatal. Back in 2015, a team of UNIMAS researchers discovered that P. knowlesi infections in Malaysian Borneo are caused by two genetically divergent subpopulations of the parasite: cluster 1 is associated with long-tailed macaque hosts, and cluster 2 is associated with pig-tailed macaque hosts.
“The crucial question is to find out whether these parasite subpopulations are changing in numbers over time,” explains Paul Cliff Simon Divis, director of the UNIMAS Malaria Research Centre.
Dr Divis used the new PCR genotyping assay to clarify different P. knowlesi infections in malaria patients.
To do this, the researchers developed a new genetic test to distinguish between and survey the two parasite subpopulations. While previously used methods, such as microsatellite genotyping and whole genome sequencing, are time consuming and expensive, the newly developed genotyping tool – a type of polymerase chain reaction (PCR) assay – is simple and efficient.
“The new test is remarkably sensitive and can detect as few as four parasites per microlitre of blood,” says Divis.
The researchers used the tool to analyse 1,204 P. knowlesi infections in patients at Hospital Kapit in Sarawak state. They found the cluster 1 parasite subpopulation, associated with long-tailed macaques, accounted for two-thirds of all cases between 2000 and 2018. This makes sense because long-tailed macaques prefer cropland, wetland and urban areas, bringing them into close proximity with humans. In contrast, pig-tailed macaques prefer more remote forests, making transmission of the cluster 2 subpopulation less likely. These findings were published in the journal Emerging Infectious Diseases.
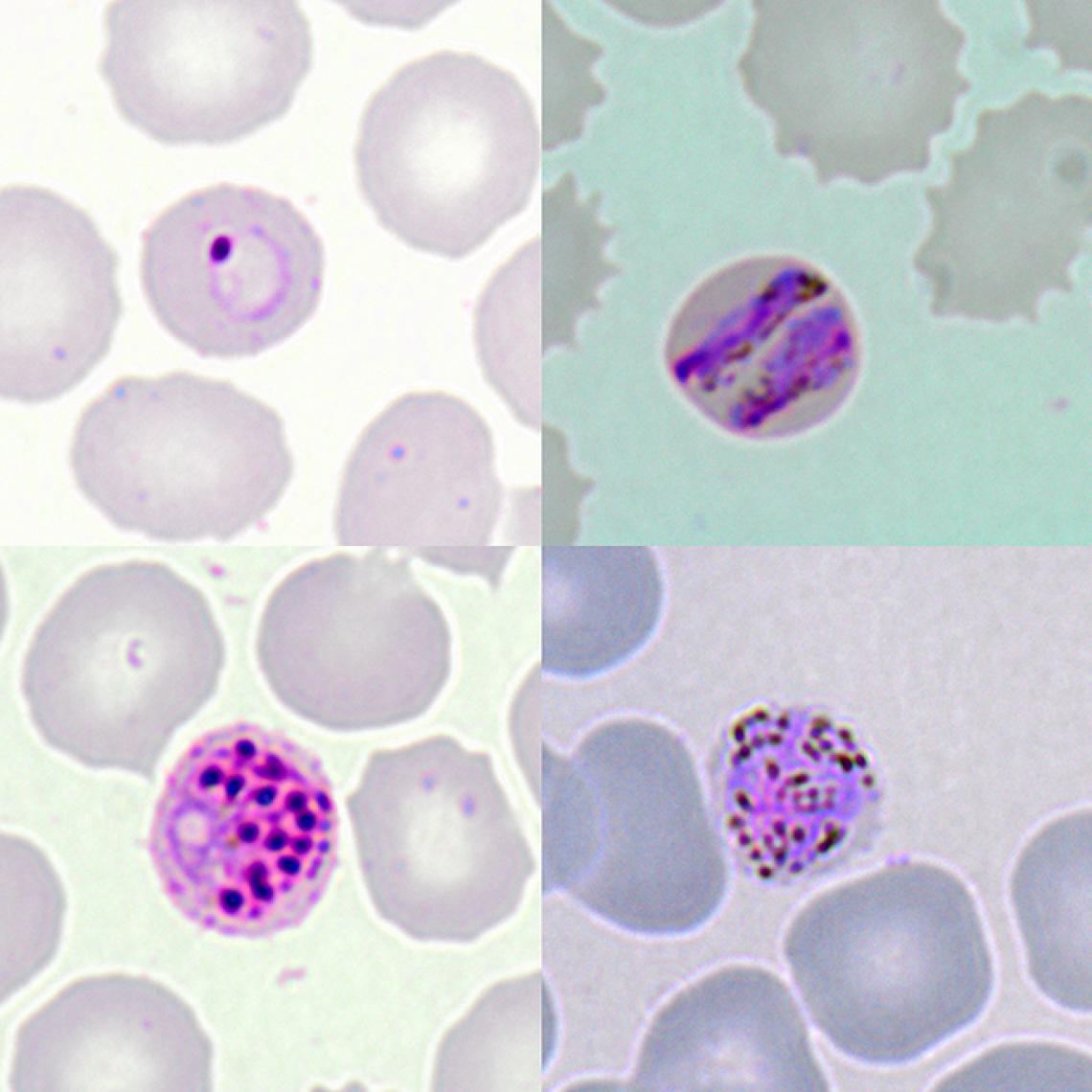
Microscopic images show the different development stages of P. knowlesi in infected red blood cells of a patient. Top Left: early ringform, Top Right: band-form trophozoite, Bottom Left: schizont, and Bottom Right: gametocyte.
The results also revealed an apparent increase in overall P. knowlesi infections between 2016 and 2018, driven mainly by rise in cluster 1 infections, which might have been caused by increased exposure to animals along with a reduction of other endemic malaria parasites.
“What’s exciting about this study is that it reveals new insights into the biological, clinical and ecological characteristics of the P. knowlesi parasite and how it infects humans,” says Divis.
Continued monitoring of the different parasite subpopulations along with tracking environmental changes, such as deforestation, may play an important part in future strategies for malaria prevention and control.
Further information
Dr Paul Cliff Simon Divis
[email protected]
Malaria Research Centre
Universiti Malaysia Sarawak (UNIMAS)